What is Drugs Technical Advisory Board?
The Drugs Technical Advisory Board advises the Central Government and the State Governments on technical matters arising out of the administration of Drugs & Cosmetics Act and carry out functions assigned to it by this Act.
Who are the members of DTAB Drugs Technical Advisory Board?
(i) the Director General of Health Services, ex officio, who shall be Chairman;
(ii) the Drugs Controller, India, ex officio;
(iii) theDirector of the Central Drugs Laboratory, Calcutta, ex officio;
(iv) the Director of the Central Research Institute, Kasauli, ex officio;
(v) the Director of Indian Veterinary Research Institute, Izatnagar, ex officio;
(vi) the President of Medical Council of India, ex officio;
(vii) the President of the Pharmacy Council of India, ex officio;
(viii) the Director of Central Drug Research Institute, Lucknow, ex officio;
(ix) two persons to be nominated by the Central Government from among persons who are in charge of drugs control in the States; one person, to be elected by the Executive Committee of the Pharmacy Council of India, from among teachers in pharmacy or pharmaceutical chemistry or pharmacognosy on the staff of an Indian university or a college affiliated thereto;
(xi) one person, to be elected by the Executive Committee of the Medical Council of India, from among teachers in medicine or therapeutics on the staff of an Indian university or a college affiliated thereto;
(xii) one person to be nominated by the Central Government from the pharmaceutical industry;
(xiii) one pharmacologist to be elected by the Governing Body of the Indian Council of Medical Research;
(xiv) one person to be elected by the Central Council of the Indian Medical Association;
(xv) one person to be elected by the Council of the Indian Pharmaceutical Association;
(xvi) two persons holding the appointment of Government Analyst under this Act, to be nominated by the Central Government.
Functions of Drugs Technical Advisory Board
1. Co-ordinate the DTAB meetings under the Chairmanship of Director General of Health Services (DGHS) to advise the Central Government and the State Governments on technical matters arising out of the administration of the Drugs and Cosmetics Act, 1940 and to carry out the other functions assigned to it by this Act.
2. Co-ordinate the DCC meetings under the Chairmanship of Drugs Controller General (India) to advise the Central Government, the State Governments and DTAB on any other matter tending to secure uniformity throughout India in the administration of the Drugs and Cosmetics Act, 1940.
3. Initiate the amendments in the Drugs and Cosmetics Rules, 1945 as per the recommendations of DTAB and co-ordinate with Ministry of Health & Family Welfare (MOHFW) for draft and final Gazette Notifications.
4. Examination, compilation and consideration of comments/suggestions/objections received with respect to draft Gazette Notifications / Public Notices / Circulars etc.
5. Co-ordinate the constitution of sub-committees recommended in DTAB and DCC meetings and further follow-up for their reports.
6. Co-ordinate the stake holders meetings with respect to amendments of Drugs and Cosmetics Rules, as per recommendations from MOHFW whenever required.
7. Prepare minutes of DTAB and DCC meetings and upload on CDSCO website for stakeholders/public reference.
8. Processing of representations/RTIs/Public Grievances with respect to Drugs and Cosmetics Act and Rules thereunder
Tenure of Drugs Technical Advisory Board Members?
The nominated and elected members of the Board shall hold office for three years, but shall be eligible for renomination and re-election:
1 [Provided that the person nominated or elected, as the case may be, under clause (ix) or clause (x) or clause (xi) or clause (xvi) of sub-section
(2) shall hold office for so long as he holds the appointment of the office by virtue of which he was nominated or elected to the Board.]
(4) The Board may, subject to the previous approval of the Central Government, make bye-laws fixing a quorum and regulating its own procedure and the conduct of all business to be transacted by it.
(5) The Board may constitute sub-committees and may appoint to such sub-committees for such periods, not exceeding three years, as it may decide, or temporarily for the consideration of particular matters, persons who are not members of the Board.
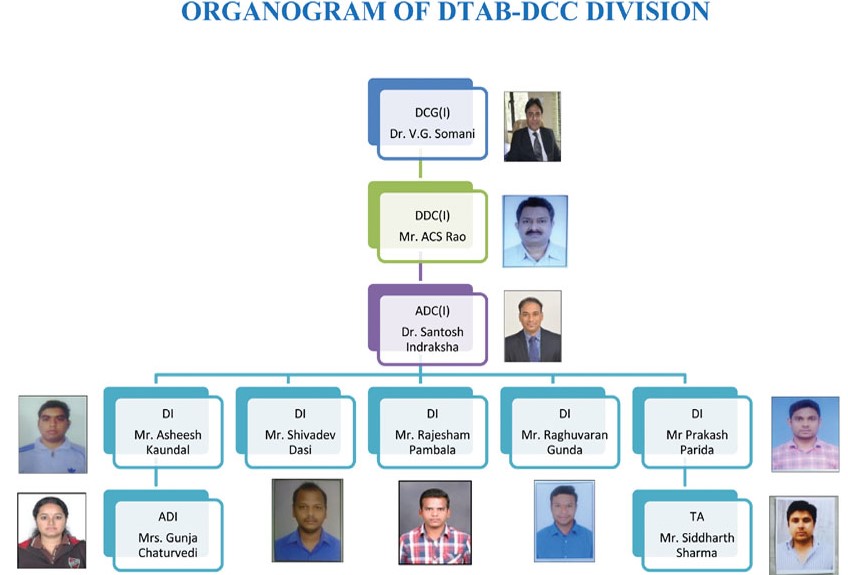
Organogram of DTAB
How DTAB Works ? Processes
1. The Chairman of the Board shall fix the date, time and place of every meeting of the Board, provided that the Board shall meet at least once every calendar year.
2. The Chairman of the Board shall, when present, preside all the meetings of the Board. If the Chairman is not present in any meeting, the members present shall select from amongst themselves a person to preside as Chairman at such meeting.
3. Seven member of the Board present in person shall constitute a quorum.
4. (i) Not less than 35 clear days’ notice of every meeting shall be given to each member of the Board, provided that the Chairman:-
(a) may call, after giving not less than 15 clear days’ notice to the members of the Board, a special meeting at any time to deal with any urgent matter requiring the attention of the Board;
(b) shall call, after giving not less than 15 clear days’ notice to the members of the Board, a special meeting within one month of the receipts of a requisition in writing signed by not less than seven members and stating the purpose, being a purpose within the scope of the Board’s functions for which they desire the meeting to be called.
(ii) The notices shall be dispatched to the latest address given to the Secretary by the members of the Board.
5. Any member desirous of moving any resolution at a meeting of the Board shall give notice, thereof in writing to the Secretary not less than 15 days before the date of such meeting, provided that in the case of a special meeting the notice will not be less than 5 days.
6. Each member of the Board shall have one vote, and if there shall be an equality of votes on any question to be decided, the Chairman shall have a second or casting vote.
7. Any business, which it may be necessary for the Board to discuss and decide except such as the Chairman may direct to be transacted by circulation among all members of the Board shall be placed at a meeting of the Board. If three or more members express, in writing, a desire that any particular subject shall be discussed at a meeting instead of being decided by circulation it shall be placed before a meeting of the Board.
8. Any resolution or report which is circulated on the direction of the Board or by the Chairman under Bye-Laws 7, and approved by a majority of the members signing shall be as binding as a resolution voted in a meeting of the Board, provided that at least nine members of the Board shall have recorded their views on the resolution in support.
9. Proceedings of each meeting, duly approved by the Chairman, shall be forwarded to the members of the Board for their approval or comments within 35 days of the date on which the meeting was held.
10. The quorum for a sub-committee appointed by the Board shall be determined at the time of the appointment of the sub-committee and shall not be less than a majority of the members appointed.
11. The Chairman and the Secretary of the Sub-committee shall be appointed by the Board at the time of the appointment of the sub-committee.
980529/2018/CDSCO-(HQ)